Bacteria
.
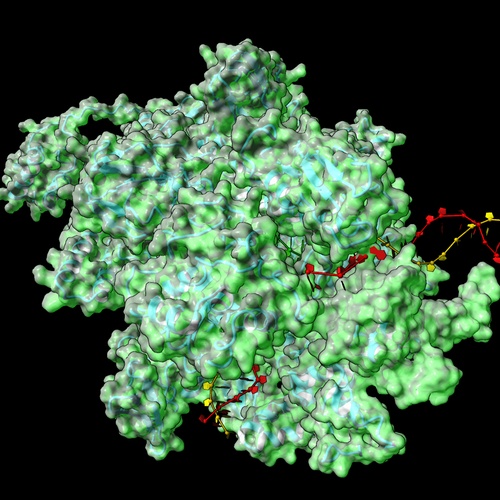
RNA polymerase as a universal damage sensor
A transcription complex is more than the static totality of its core subunits and factors. Rather, it is a dynamic ensemble comprised of different interacting molecular partners that perpetually associate and dissociate from the RNA polymerase. Our lab studies the dynamic interactome of bacterial RNA polymerases as they react to genotoxic and proteotoxic stresses and directly couple transcription to other major cellular processes, such as translation and DNA repair. We use a combination of bacterial genetics, in-vitro biochemistry, in-house developed next generation sequencing (NGS) protocols, high-resolution structural proteomics, and cryo-electron microscopy to characterize the relevant molecular complexes.
Relevant publications
- Rates and mechanisms of bacterial mutagenesis from maximum-depth sequencing. Nature. 30 Jun 2016. 534(7609):693-6
- ppGpp couples transcription to DNA repair in E. coli. Science. 20 May 2016. 352(6288):993-6
- UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 16 Jan 2014. 505(7483):372-7
H2S defense system
Most bacteria (and other organisms) enzymatically produce hydrogen sulfide (H2S) from L-cysteine and its derivatives. Although potentially toxic, H2S at physiological concentrations acts as an important signaling molecule and effectively protects cells from oxidative stress. The H2S defense system contributes substantially to antibiotic resistance of bacterial pathogens, and protects them against immune attack. Our lab studies the enzymology and molecular biology of the H2S-mediated stress response in Gram-positive and Gram-negative pathogenic bacteria. We also screen large chemical libraries for small molecule inhibitors of H2S biosynthesis, characterize the biochemical properties of top hits, and derivatize them to improve their pharmacokinetic parameters. Our goal is to identify promising candidates that potentiate the activity of many classes of antibiotics and even prove useful for treating some pathologies of human cells.
Relevant publications
- Inhibitors of bacterial H2S biogenesis targeting antibiotic resistance and tolerance. Science. 11 Jun 2021. 372(6547):1169-1175
- H2S: a Bacterial Defense Mechanism Against the Host Immune Response. Infect Immun. 15 Oct 2018. epub ahead of print
- Mechanism of H2S-mediated protection against oxidative stress in Escherichia coli. Proc Natl Acad Sci U S A. 06 Jun 2017. 114(23):6022-6027
- H2S: a universal defense against antibiotics in bacteria. Science. 18 Nov 2011. 334(6058):986-90